Saving and Archiving Data
- You should be logged in and running xwinnmr under your user id.
- All data is saved to x/data/"user id"/nmr/
- I archive all data to CD on the first of each month.
- You can FTP data from the SGI to the nmr PC and burn to CD. See me and I can help you do this.
General Procedures for observing nuclei other than 1H
- Insert sample and cable to the following diagram for the BBO (When using the TBI change the cable from 13C to BB on the probe head):
- Before inserting the probe tune and match to the rough values on the card (except when observing 2H, the X channel needs to be turned away from 2H to start).
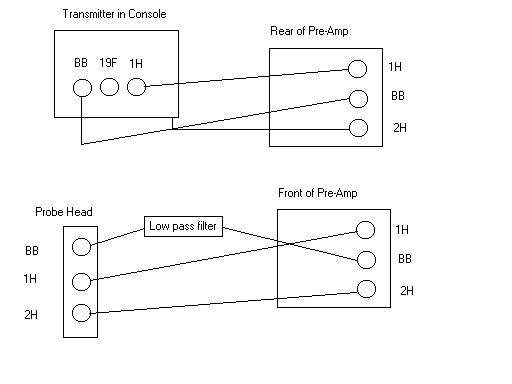
- Read a standard parameter file for the nuclei you want to observe:
| Nucleus to observe | Parameter set | Comments |
| 13C without 1H coupling | C13CPD | sensitivity limited |
| 13C attached proton test | C13APT | nice experiment |
| 17O | O17 | sensitivity limited |
| 15N | N15 | sensitivity limited |
| 15N with 1H decoupling
|
N15
|
better sensitivity. For 1H decoupling change the pulse program to zgdc and make sure in edsp the F2 is set to 1H. |
| 31P | P31 | sensitivity limited |
| 31P with 1H decoupling | P31CPD | set pl12 and pl13 to 20 , better sensitivity |
| 1H with 31P decoupling | PRO31PDEC | optimization is questionable |
| 2H with and without 1H decoupling
|
DEUTERIUM
|
Tune, match, lock and shim for a normal 1H spectra. For acquisition recall the parameter set. Change locknuc to 2H and change your solvent then turn locknuc off. On the BSMS; no sweep or lock, lock power to -60, lock gain to 75, no filters on X channel. For 1H decoupling change the pulse program to zgdc and make sure in edsp the X nuc is going to the 2H preamp and F2 is set to 1H. |
- Read shims for your solvent.
- Lock on your solvent.
- Tune and match with the sliders (and knobs if also interested in 1H)
- Run your experiment
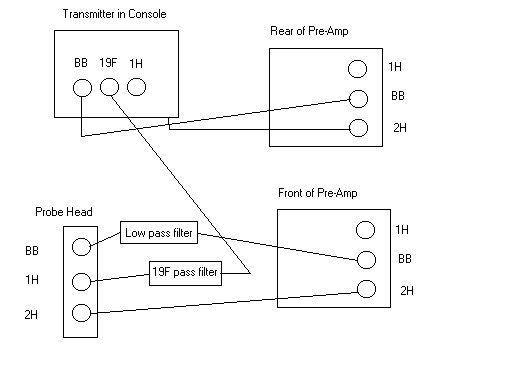
Observe 13C and decouple 19F
- Using the BBO probe obtain a normal 13C spectrum and set the SW and O1 to region of interest
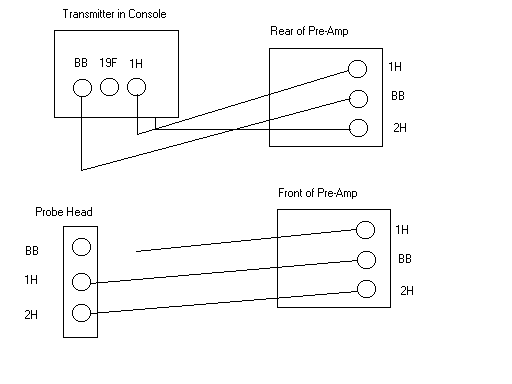
- Next, you must detune the 1H channel to 19F
- Remove the 1H cable from the probe head
- Remove the BB cable from the probe head and connect it to 1H input of the probe head.
- See figure below for cables:
- Next you will collect the 19F spectrum. Type rpar F19 all then type ii
- Type a and then wobb
- Change the sweep width to 20MHz by typing wbsh and then entering 20MHz.
- Tune and match the signal on the 1H cannel. Tune first then match. You may have to open the sweep width more to see the dip.
- Type p1 5
- Type pl1 0
- Type pulprog and confirm the pulse program is zgflqn.
- Type eda, change the solvent and prosol to true.
- Type edsp and make sure the connections are as shown below:
- Type rga then zg. Process and calibrate the 19F spectrum.
- Display the region of interest on the screen and click on the sw-sfo1 button. Write down the SW and O1 frequency.
- Now you will recable and collect the 19F decoupled 13C spectrum.
- Type iexpno to start a new experiment and type rpar C13CPD all.
- Type eda , set the solvent and prosol to true.
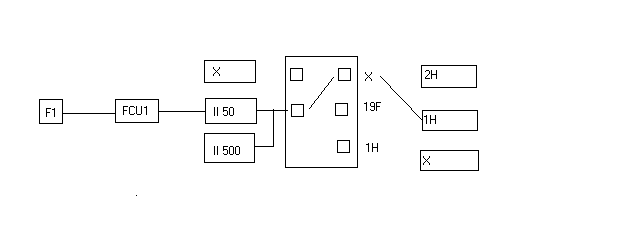
- Click on NUCLEI edit and change NUC2 to 19F and make the following connections:
- Check 19F as the preferred output for 19F.
- Click on SAVE and OK the message " Warning: connection to preamplifier is missing"
- In the eda window, change O2 to the value of O1 you found in the 19F spectrum.
- Change CPDPRG2 to the following depending upon the SW to be decoupled in the 19F spectrum:
- Use the following parameters in the PCPD and PL arrays for the particular CPDPRG2 used:
- You may have to optimize PL (12) for the best results.
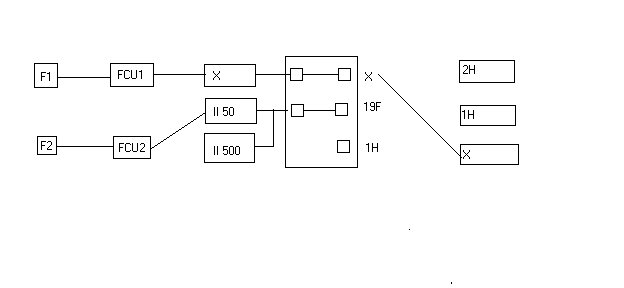
- Click on SAVE and then recable according to the following diagram:
- Type rga and then zg.
- When finished recable as show below:
|
|
|
| Less than 10,000 Hz | waltz16 |
| Between 10,000 and 30,000 Hz | garp |
| More than 30,000 Hz | p5m4 |
|
|
|
|
|
| waltz16 | 90 usec | 20 db | 20 db |
| garp | 64 usec | 14 db | 14 db |

DPFGSE -1D NOESY(This experiment provides nOe information on a peak by peak basis. Each 1D scans takes about 3 to 6 minutes depending upon number of scans. This is much faster than a 7 to 12 hr 2D-NOESY experiment and should be considered for small molecules.)
- Take a reference proton spectra without spinning. Use the minimum number of points (td) as you can tolerate. Set o1 to the middle of the spectra and narrow the sw to encompass all the peaks. Note your value of o1 since all experiment will need this reference value. Write down the Hz value for each peak you want to investigate.
- Increment the experiment with iexpno and rpar sel-1D-noe all
- Type eda and change your solvent and then change prosol to true.
- Verify the following:
| Parameter | Comment |
| o1 | same as reference proton |
| sw | same as reference proton |
| td | same as reference proton |
| si | same as reference proton |
| ns | 64 to 128 is ok |
| ds | 4 |
- Type ased and make sure the following parameters are correct:
| Parameter | Value |
| spoff1 | (Hz value of peak - o1 value) can be negative |
| sp1 | 70 db |
| p12 | 100 msec (the 180 deg shaped pulse, the 90 is 50 ms @ 70 db) |
| d8 | mixing time (700 ms for small molecules) |
| d20 | d8/2 -1.5 msec |
- Type zg, when finished type ef and phase so the peak of interest is negative.
- Use xwinplot to do a stacked plot.
Water Suppression Parameter Sets
| Parameter Sets | Comments |
| jump-return-drug-dna | must set the o1p to water |
| dpfg-water-supression | must set the o1p to water (works the best) |
| watergate.mww | must set the o1p to water |
Kinetics Array
- Set up experiment (use temperature control) and narrow sweep width to the peak(s) of interest.
- Define the plot region.
- Find the amount of time it takes for 1 experiment by typing expt.
- Set the time between experiments by calculating calculate d1: (d1 = Desired time increment - time for one experiment determined from expt command)
- Type paropt and enter.
- Make d1 the parameter to change, initial value is the value you calculated two steps above, increment will be 0 and the number of experiments should span 4 to 5 half lives.
- The spectra are displayed horizontally and can be integrated.
- If you get a message that the file size is too big try decreasing td. You will also need to make si = td/2. Sometimes making a new file with the edc command will allow the experiment to run.
1H - 31P HMBC
- Run you 1H spectra on the region of interest and record the sw, o1p and file name.
- Run the 31P spectra with 1H decoupling and record the sw, o1p and file name.
- Run the 31P spectra without 1H decoupling.
- The HMBC parameter set is called H_P_HMBC
- Change the following parameters in the order that are encountered below:
| Parameter | Value |
| prosol | true |
| solvent | your solvent |
Gradient Selected COSY (gCOSY)
- Obtain a normal 1H NMR spectrum (see previous section)
- Expand your spectrum to the region of interest
- Click on DP1 (lower left) to define the plot region and then click on SF-SFO1 to define the sweep width (SW) and O1 (off set)
- Type sw and write down the value. Press enter to go back to the command line and type o1 and write down its value.
- Type rga then zg to Collect and process a proton spectrum to display on the F2 and F1 dimensions (write down the file name and spectrum number)
- Stop the sample from spinning and shim.
- Type iexpno to increment the experiment number.
- Type rpar wfu.4min.gCOSY all to copy all the gCOSY parameters
- Type eda and change the existing values to those listed below:
| Parameter |
|
|
|
Notes |
| TD | 2048 | 128 | Use 4k and 2k for better resolution of crowded spectra | |
| NS | 1 | |||
| DS | 8 | |||
| SW | determined value | determined value | From 1H spectra | |
| O1 | determined value | determined value | From 1H spectra | |
| Solvent | Choose your solvent | |||
| PROSOL | True | Must be set when a new parameter set is recalled |
- Click on Save
- Type expt to see how much time the experiment will take. You can change the values of TD to alter the desired S/N or resolution. Those listed are the minimum and will only take 4 to 5 minutes. When TD is set to 4096 and 2048 it will take about 1 hr and 20 minutes and gives better resolution.
- Type rga to reset the gain
- Type zg to start the experiment
- Type edg to edit the graphics menus (this can be done while the experiment is running)
- Click on EDPROJ1 and make these changes:
| Parameter | Value |
| PF1USER | your login name |
| PF1NAME | exact name of the 1H NMR spectrum you initially ran |
| PF1EXP | experiment number of the 1H NMR spectrum you initially ran |
- Click on Save
- Click on EDPROJ2 and make these changes
| Parameter | Value |
| PF2USER | your login name |
| PF2NAME | exact name of the 1H NMR spectrum you initially ran |
| PF2EXP | experiment number of the 1H NMR spectrum you initially ran |
- repeat the previous step and save all changes. (all the parameters names with a 1 will now have a 2 in the name)
- Type xfb when the experiment is done.
- See the section on 2D Spectra Manipulation to plot and expand regions of your 2D spectrum.
Gradient HMQC (g-HMQC)
- Collect and process a proton spectrum of the region in interest (write down the file name, spectrum number, SW and O1)
- Collect and process a proton decoupled carbon spectra (write down the file name, spectrum number, SW and O1)
- Stop the sample from spinning and shim.
- Type iexpno to increment the experiment number.
- Type rpar wfu.birdHMQC all
- Type eda and change the existing values to those listed below:
|
|
|
|
|
|
| PULPROG | invbst | |||
| TD | 1k | 128 | ||
| NS | 8 | |||
| DS | 16 | |||
| NDO | 2 | |||
| NUCLEI | EDIT and change NUC2 from off to 13C | EDIT and choose 13C | ||
| SW | value from 1H spectra | value from 13C spectra or use 230 if no carbon has been run | ||
| O1 | value from 1H spectra | |||
| O2P | value from 13C spectra or use 1/2 of SW for F1 if no carbon has been run | |||
| SOLVENT | Chose your solvent | |||
| PROSOL | True |
- Click on Save
- Verify the following values by typing in the parameter and pressing enter. Press enter again and then go to next parameter.
| Parameter | Value |
| PL1 | Pulse and Delay Appendix |
| PL2 | Pulse and Delay Appendix |
| P1 | Pulse and Delay Appendix |
| P2 | Pulse and Delay Appendix |
| P3 | Pulse and Delay Appendix |
| P4 | Pulse and Delay Appendix |
| D1 | 1.5* average T1 (1.5 s should be OK) |
| CNST | 145 |
| D2 | 0.00345 s |
| CPDPRG2 | garp |
| D7 | see section on Optimizing D7 |
- Optimizing D7: Type D7 and enter 0.400 s. Type a to see the acquisition window. Type gs and you should see the FID. Change D7 to 0.1, wait a few seconds and note the amplitude of the FID. Increase D7 by 0.1 increments. Look for the smallest FID amplitude and note the value of D7.
- Type expt to see how much time the experiment will take. These parameters will take about 1.5 hours.
- Type rga to reset the gain
- Type zg to start the experiment
- Type edg to edit the graphics menus (this can be done while the experiment is running)
- Click on EDPROJ1 make these changes:
| Parameter | Value |
| PF1EXT | Click and choose Positive |
Parameters when there is a stored 13C spectrum:
| Parameter | Value |
| PF1USER | your login name |
| PF1NAME | exact name of the 13C NMR spectrum you initially ran |
| PF1EXP | experiment number of the 13C NMR spectrum you initially ran |
- Click on Save
- Click on EDPROJ2 make these changes:
| Parameter | Value |
| PF2USER | your login name |
| PF2NAME | exact name of the 1H NMR spectrum you initially ran |
| PF2EXP | experiment number of the 1H NMR spectrum you initially ran |
- Type xfb when the experiment is done
- Type prow to phase the first row of data. Phase the spectra manually and click on Return and choose "Save as 2D and return"
- Type xau calcphinv then enter xfb.
- Set the contour level with the *2 and /2 buttons.
- See the section on Phasing 2D Spectra if phasing is needed.
- See the section on Manipulating and Plotting 2D Spectra your 2D to plot regions of interest.
Gradient NOESY (g-NOESY)
- Obtain a normal 1H NMR spectrum (see previous section)
- Expand your spectrum to the region of interest
- Click on DP1 (lower left) to define the plot region and then click on SF-SFO1 to define the sweep width (SW) and O1 (off set)
- Type sw and write down the value. Press enter to go back to the command line and type o1 and write down its value.
- Type rga then zg to Collect and process a proton spectrum to display on the F2 and F1 dimensions (write down the file name and spectrum number)
- Stop the sample from spinning and shim.
- Type iexpno to increment the experiment number.
- Type rpar wfu.gNOESY all.
- Type eda and change or verify the existing values to those listed below: (these can entered through the command line using small case letters)
|
|
|
|
|
|
| PULPROG | noesygpst | |||
| PL1 | Pulse and Delay Appendix | |||
| DO | 3 usec | |||
| D8 | 250 msec | just a starting value | ||
| D16 | 100 usec | |||
| TD2 | 2k | |||
| SW2 | from previous 1H experiment | |||
| O1 | from previous 1H experiment | |||
| NS | 2 | |||
| P1 | Pulse and Delay Appendix | |||
| P2 | Pulse and Delay Appendix | |||
| P16 | 1 msec | |||
| D1 | 2 sec | |||
| D11 | 30 msec | |||
| D20 | D8*0.5 - P16 -D16 | |||
| ND0 | 1 | |||
| TD1 | 256 | |||
| SW1 | from previous 1H experiment | |||
| L3 | use default | |||
| DS | 16 | |||
| gpnam1 | SINE.1000 | |||
| gpnam2 | SINE.1000 | |||
| INO | 1/(SW1) | |||
| gpz1 | 40% | |||
| gpz2 | -40% |
- Type rga then zg
- Type edp to change the processing parameters to:
|
|
|
|
| SI(F2) | 512 | |
| WDW(F2) | EM | |
| LB(F2) | 2 | |
| PH-mod(F2) | pk | |
| PHC1(F2) | ||
| MC2 | ||
| SI(F1) | ||
| WDW(F1) | ||
| SSB(F1) | ||
| PH-mod(F1) | ||
| PHC1(F1) |
- Phase the rows and columns as presented in the gs-COSY experiment.
Selective Pulse Lengths and Powers:
- 1H 90o 50 ms @70db
- 13C 90o ____ @ ____