Research Interests
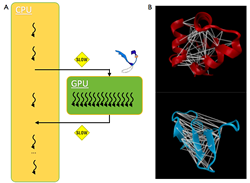
Jump to: OverviewOur interdisciplinary research group interests encompass biophysics and computer science. As such, our group is represented with current and previous undergraduate and graduate students with physics, computer science, chemistry, biology, and mathematics backgrounds. We are interested in the theoretical and computational studies and methods development of biomoleular coarse-grained and atomistic molecular dynamics (MD) simulations in collaboration with experimental groups. The inherent computational complexity of MD simulations, however, require cutting-edge high performance computing to perform simulations of biologically relevant sized systems for timescales that correspond to biological functions. One of our primary focuses is to develop MD simulation approaches using graphics processing units (GPUs), which are ideally suited for parallel algorithms such as those in MD simulations. Below are brief descriptions of a few ongoing research projects. GPU-Optimized Coarse-Grained MD Simulations
Understanding protein and RNA biomolecular folding and assembly processes have important applications because misfolding is associated with diseases like Alzheimer’s and Parkinson’s. However, simulating biologically relevant sized biomolecules on timescales that correspond to biological functions is an extraordinary challenge due to bottlenecks that are mainly involved in pairwise force calculations. Recent studies have demonstrated that GPUs, which were originally designed for rendering images, can be repurposed for parallel algorithms such as those in MD simulations. Our work is currently focused on developing new algorithms and features for our coarse-grained MD simulations that is specifically optimized for the GPU architecture, and it is funded by the National Science Foundation and NVIDIA, Inc. RNA Folding Mechanisms: tRNA
The RNA folding problem has become increasingly appreciated because RNA molecules play critical roles in a variety of cellular functions. In fact, in mice and human genomes, the percentage of the transcribed non-coding sequences exceed 90%, and we are just beginning to understand the functional importance of the rest of the non-coding RNA molecules. Although the non-coding roles of transfer RNA (tRNA) and ribosomal RNA (rRNA) have been well known for quite some time, but it was the discovery of ribozymes (RNA enzymes) that sparked an intense search for the functional roles of the rest of the non-coding RNA molecules. We now know that non-coding RNA molecules’ functions include catalysis, replication, translational regulation, and ligand binding, just to name a few. The changes in the expression level of non-coding RNA molecules have been associated with cancer, neurological diseases, and other disorders, suggesting crucial roles for non-coding RNA in cellular processes when they are functioning properly. We focus on tRNA molecules, which are known to fold in distinct mechanisms despite their structural similarities. Recent work using coarse-grained and atomistic MD simulations strongly suggests that the global tRNA folding mechanisms, from both thermodynamics and kinetic viewpoints, are very complex, oftentimes involving parallel folding and backtracking mechanisms. Protein-Nanoparticle Interactions: Biocorona Formation
With the advancement of nanotoxicology and nanomedicine, it has been realized that nanoparticles interact readily with biomolecular species and other chemical and organic matter. Owing to their small size, large surface area-to-volume ratio, and versatile physicochemical properties, nanoparticles readily enter cells and organelles, leading to the possibilities of new medicine (nanomedicine) or biological hazard (nanotoxicity). The field of the environmental health and safety of nanotechnology, or NanoEHS, is currently lacking significant molecular-resolution data. A key concept emerging in the fields of nanomedicine and nanotoxicity is the idea of protein-nanoparticle interactions that result in a dynamic layer of proteins and other biomolecules, or “corona”, that adsorbed onto a nanoparticle surface. In this view, nanoparticles entering any biological fluid will be covered by a corona of biomolecules and disrupt the native conformation and, therefore, compromise the function of the protein. Recently, we developed a novel GPU-optimized coarse-grained MD simulation methodology for the study of biocorona formation, a first in the field. This work is in collaboration with the group of Pu-Chun Ke (Clemson University), and it is funded by the National Science Foundation. Protein/RNA Complexes: tRNA:Aminoacyl-tRNA Synthetases
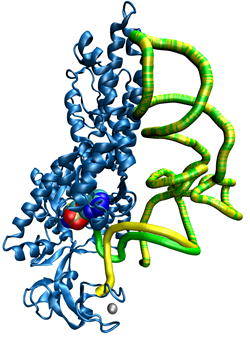
Aminoacyl-tRNA synthetases (AARSs) are enzymes that are responsible for chemically connecting an amino acid and its associated tRNA through a process called aminoacylation. The activated tRNAs are transported to the ribosome, where the amino acids are strung together to form proteins. In order for the AARSs to complete its catalytic function, it has to recognize the anticodon and aminoacylation region at the same time.Because of its fundamental role in biology and its versatility, AARSs have been linked to cancer, neuronal pathologies, autoimmune disorders, and disrupted metabolic pathways, which may all be due to heritable mutations. We hope that our work will contribute towards developing different treatment methods or new drugs. Our recent work is focused on performing long-timescale coarse-grained and atomistic MD simulations of a refined modeled MetRS:tRNAMet complex, the AARS and tRNA that corresponds to the amino acid methionine. One of our primary focuses is the CP domain, which is an editing domain in other AARSs, but its biological function in MetRS is unknown. We hypothesize a fly-casting type mechanism that recognizes and facilities the binding of tRNAMet through an allosteric communication and signaling network. This work is in collaboration with the group of Rebecca Alexander (Wake Forest University, Department of Chemistry), and it is funded by the Wake Forest University Center for Molecular Communication and Signaling. DNA Dynamics: G-Quadruplexes
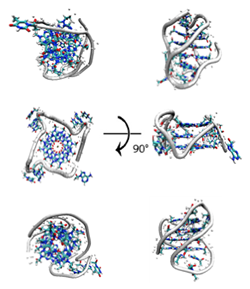
Telomeres are the chromosomal regions that are removed during replication and replaced by the telomerase enzyme. In normal cells, the activity of the enzyme is low, and the telomeres get shorter with each successive cell division, resulting in eventual apoptosis. However, one of the hallmarks of cancer cells is the overexpression of the telomerase enzyme that results in telomere lengthening, resulting in cellular over-proliferation or cancer. These telomere regions contain guanine-rich ends that are capable of forming structures known as G-quadruplexes. They consists of two strands with two tetrads each forming a quadruplex or four small strands with one tetrad each that combine to form a quadruplex. These quadruplexes have a great deal of structural polymorphisms such that they can adopt one of many different topologies with different backbone and loop representations. Our recent work is focused on performing long-timescale coarse-grained and atomistic MD simulations of putative ribosomal DNA G-quadruplexes by threading sequences to known topologies. The DNA G-quaduplexes are potential targets of intercalating drugs that inhibit telomerase-catalyzed strand elongation by stabilizing the G-quadruplex. This work is in collaboration with the group of Uli Bierbach (Wake Forest University, Department of Chemistry). |