The Alexander Lab
Current Research Projects
|
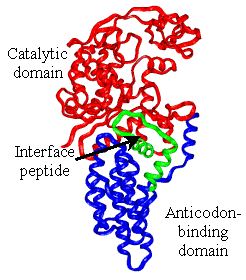
Our lab is investigating the long-range communication in E. coli methionyl-tRNA synthetase (MetRS), an AARS that requires anticodon binding for efficient catalysis yet also aminoacylates a small tRNA mimic lacking an anticodon, making it a good model to study mechanisms of long-range communication in proteins. We have used computational and experimental techniques to address how long-range communication occurs in MetRS. The computational project is in collaboration with Dr. Jacque Fetrow (WFU Physics and Computer Science Departments), and has demonstrated that functional regions of the protein exhibit correlated motions and that there are low energy barriers to achieve the alternate conformations observed by x-ray crystallography. We have made mutations at residues predicted from structural comparisons and computational methods to be important in long-range communication, and preliminary results indicate that some of these mutations do impact tRNA aminoacylation efficiency. |
|
As we investigate the contribution of protein local structure to RNA recognition and enzyme catalysis, we have swapped amino acid residues from the stem-contact fold domain of glutaminyl-tRNA synthetase (GlnRS) into the structurally similar domain of MetRS. This structural motif has been proposed to interact with the extreme inner corner of each enzyme’s cognate tRNA to globally orient the nucleic acid on the protein surface. Our hypothesis is that some portions of this structurally conserved motif can be swapped between proteins with minimal effect on tRNA binding and aminoacylation, while other residues may participate in essential unique interactions with each enzyme’s active site and substitution will not be tolerated. Analysis of the domain swapped constructs and other mutants already constructed by undergraduate and graduate students in the lab will allow continued evaluation of the network of interactions necessary for efficient catalysis. Funding for these projects is from the National Science Foundation and the National Foundation for Cancer Research. |
The side-effects of accuracy control
The aminoacyl-tRNA synthetases are responsible for maintaining the fidelity of the genetic code by accurately attaching amino acids with their matching tRNAs. In some cases, synthetases actively prevent mis-incorporation of amino acids into proteins by hydrolyzing noncognate aminoacyl adenylates or aminoacyl-tRNAs. One of the amino acids that may be processed this way by MetRS is homocysteine, which lacks the terminal methyl group of methionine. For some time there has been clinical evidence that high serum homocysteine levels are correlated with diseases such as arthrosclerosis and lupus. We are interested in investigating the link between MetRS and the chemical intermediates formed when homocysteine is edited out of the biosynthetic pathway. We are working with investigators at the Scripps Research Institute in California to study the role of MetRS in human disease.
The impact of minor-groove DNA adducts on cellular processes
As an organism’s genes are transcribed into RNA for protein biosynthesis, protein transcriptional factors regulate which genes are transcribed by binding DNA sequence elements termed promoters. Binding typically involves hydrogen bonding interactions between solvent-exposed amino acids on the transcription factor and base pair atomic groups in the DNA’s major groove. We have been collaborating with Dr. Uli Bierbach (WFU Chemistry Department) to study the molecular effects of treating DNA with the novel platinum-containing compound PT-ACRAMTU. We have confirmed, using restriction enzyme analyses and transcriptional footprinting, that PT-ACRAMTU modifies adenine bases in TpA and GpA steps. Covalent modification inhibits T7 RNA polymerase from transcribing DNA, which may point to a mechanism for the compound’s demonstrated activity against cancer cells. Electrophoretic mobility shift assays with the TATA-binding protein (TBP, a minor groove binding protein) demonstrated that adducts formed at the N3 position of adenine, in the DNA minor groove, are most effective at blocking TBP binding and probably represent the physiologically relevant adduct. We will continue to probe the molecular impact of DNA damage using the tools of biochemistry and molecular biology, and are expanding our efforts to investigate the affinity and sequence specificity of PT-ACRAMTU for RNA. Funding for this project is from the National Foundation for Cancer Research.
Characterizing a cold-induced RNA helicase
Cellular information transfer is highly dynamic, with proteins processing along DNA during transcription and the ribosome traversing a messenger RNA during protein synthesis. At reduced temperatures, enhanced stability of RNA structure may prevent ribosome assembly or translation of a message. We are working with Dr. Pamela Jones at Winston-Salem State University to characterize the activity of E. coli CsdA, a protein that is produced by bacteria in response to low temperature growth conditions. CsdA has all the canonical sequence motifs attributed to the DEAD-box helicases, which are ATP-dependent RNA helicases. We have determined that CsdA indeed unwinds certain RNA substrates in the presence of ATP. The DEAD (Asp-Glu-Ala-Asp) sequence of CsdA is essential for this function. These results parallel those observed in vivo by Dr. Jones. The experimental groundwork for this project, including protein overexpression strategies and assay development, was established by a very talented WFU undergraduate working in my laboratory. We are continuing to investigate the catalytic activity and substrate specificity for this helicase. One question regarding ATP-dependent RNA helicases in general is how the free energy of ATP hydrolysis is mechanically coupled to RNA unwinding. Experiments will focus on the temperature dependence of ATP hydrolysis and RNA unwinding compared to the thermal stability of various RNA substrates. This project is funded by an NIH Research Infrastructure in Minority Institutions (RIMI) Grant to Winston-Salem State University. |
|
Rebecca W. Alexander
Department of Chemistry
Wake Forest University
Winston-Salem, NC 27109
alexanr@wfu.edu
336-758-5568 (phone)
336-758-4656 (fax)